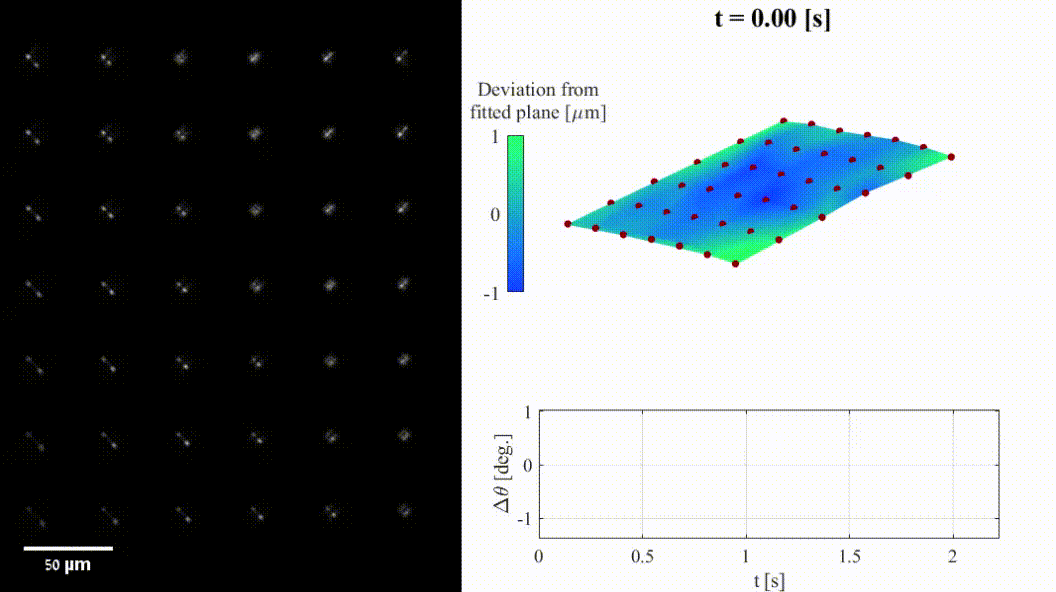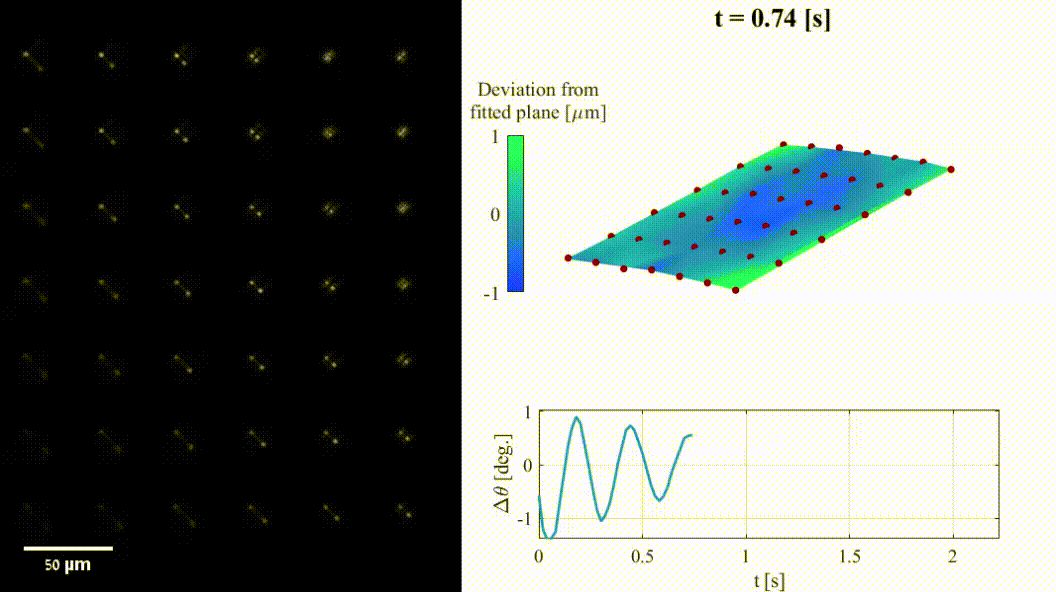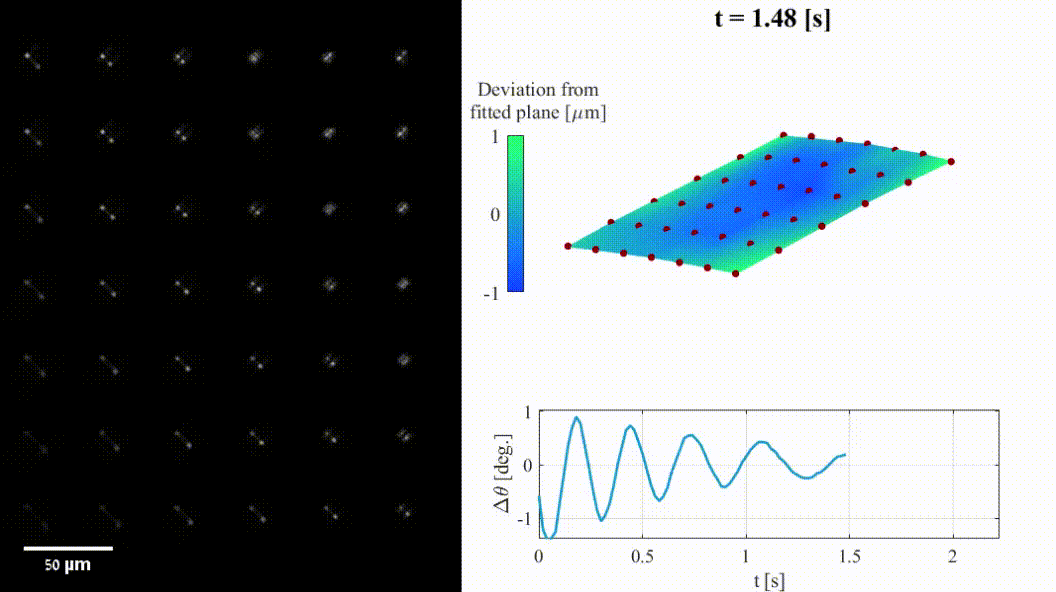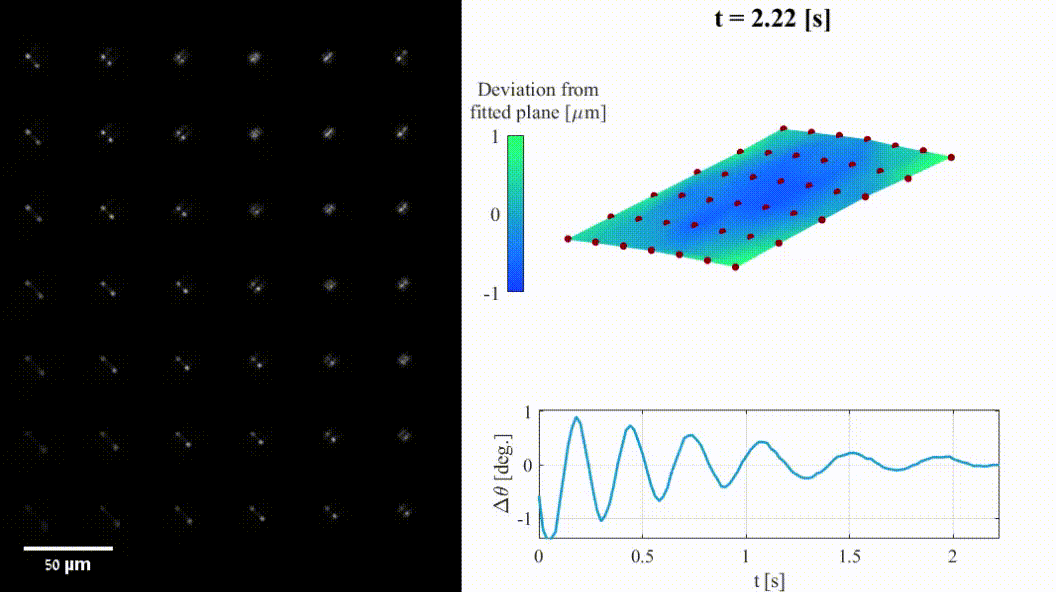
 The shape of a surface, i.e. its topography, influences many functional properties of a material; hence, characterization is critical in a wide variety of applications. Two notable challenges are profiling temporally changing structures, which requires high-speed acquisition, and capturing geometries with large axial steps. Here we leverage point-spread-function (PSF) engineering for scan-free, dynamic, micro-surface profiling. The presented method is robust to axial steps and acquires full fields of view at camera-limited framerates. We present two approaches for surface profiling implementation:
The shape of a surface, i.e. its topography, influences many functional properties of a material; hence, characterization is critical in a wide variety of applications. Two notable challenges are profiling temporally changing structures, which requires high-speed acquisition, and capturing geometries with large axial steps. Here we leverage point-spread-function (PSF) engineering for scan-free, dynamic, micro-surface profiling. The presented method is robust to axial steps and acquires full fields of view at camera-limited framerates. We present two approaches for surface profiling implementation:
- Fluorescence-based: using fluorescent emitters scattered on a surface of a 3D dynamic object (inflatable membrane in this example).
- Label-free: using pattern of illumination spots projected onto a reflective sample (tilting mirror in this example).
Both implementations demonstrate the applicability to a variety of sample geometries and surface types.
For more details:
R. Gordon-Soffer, L. E. Weiss, R. Eshel, B. Ferdman, E. Nehme, M. Bercovici, and Y. Shechtman, “Microscopic scan-free surface profiling over extended axial ranges by point-spread-function engineering“, Science Advances 6, 44 (2020).

Localization microscopy is an imaging technique in which the positions of individual point emitters (e.g. fluorescent molecules) are precisely determined from their images. Localization in 3D can be performed by modifying the image that a point-source creates on the camera, namely, the point-spread function (PSF). The PSF is engineered to vary distinctively with emitter depth, using additional optical elements. However, localizing multiple adjacent emitters in 3D poses a significant algorithmic challenge, due to the lateral overlap of their PSFs. In our latest work, DeepSTORM3D, we presented two different applications of CNNs in dense 3D localization microscopy:
- Learning an efficient 3D localization CNN for a given PSF entirely in silico (Tetrapod in this example)
- Learning an optimized PSF for high density localization via end-to-end optimization

For more details:
Nehme, E., Freedman, D., Gordon, R. et al. “DeepSTORM3D: dense 3D localization microscopy and PSF design by deep learning“. Nature Methods (2020).
 Even at the single-cell level, biological systems are both complex and heterogeneous. To understand such systems, it is essential to sample the full distribution of behaviors in a population of cells by recording many measurements. Further complicating the problem: many intracellular events of great interest occur over extremely small spatial scales; this necessitates high-resolution, 3D microscopy. In conventional microscopes, there is an inherent trade off between throughput and spatial detail that makes it challenging to achieve both of these aims concurrently. Here, we have demonstrated that by merging two technologies: point-spread-function engineering and imaging flow cytometry, we can attain excellent spatial detail with extremely high sample throughput! Essential to our approach is calibrating the imaging system. This is accomplished with a novel method that analyzes the statistical distributions of tiny fluorescent beads that are imaged alongside cells suspended in media. This is represented in the attached graphic, where the images of randomly positioned objects on the left are analyzed collectively to produce the shape-to-depth calibration on the right. This calibration is then applied to the images of fluorescently labeled positions within cells.
Even at the single-cell level, biological systems are both complex and heterogeneous. To understand such systems, it is essential to sample the full distribution of behaviors in a population of cells by recording many measurements. Further complicating the problem: many intracellular events of great interest occur over extremely small spatial scales; this necessitates high-resolution, 3D microscopy. In conventional microscopes, there is an inherent trade off between throughput and spatial detail that makes it challenging to achieve both of these aims concurrently. Here, we have demonstrated that by merging two technologies: point-spread-function engineering and imaging flow cytometry, we can attain excellent spatial detail with extremely high sample throughput! Essential to our approach is calibrating the imaging system. This is accomplished with a novel method that analyzes the statistical distributions of tiny fluorescent beads that are imaged alongside cells suspended in media. This is represented in the attached graphic, where the images of randomly positioned objects on the left are analyzed collectively to produce the shape-to-depth calibration on the right. This calibration is then applied to the images of fluorescently labeled positions within cells.
For more details:
L. E. Weiss, Y. Shalev Ezra, S. Goldberg, B. Ferdman, O. Adir, A. Schroeder, O. Alalouf, Y. Shechtman. “Three-dimensional localization microscopy in live flowing cells” Nature nanotechnology (2020).
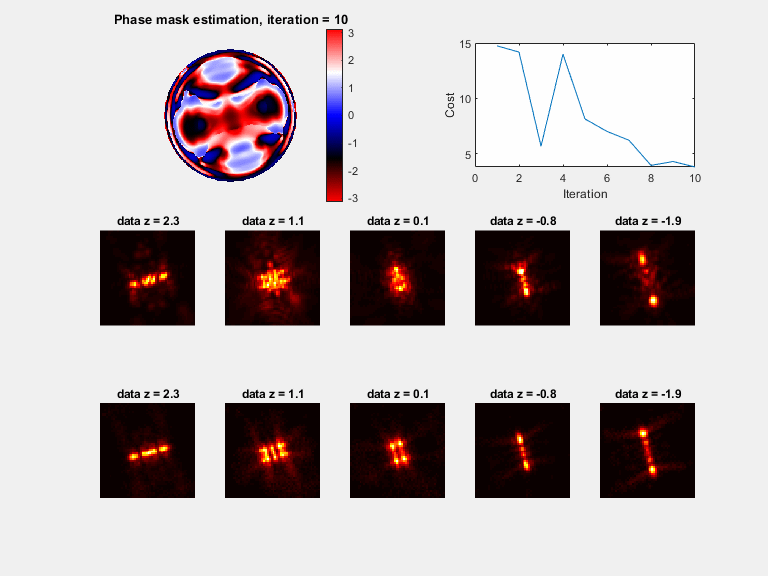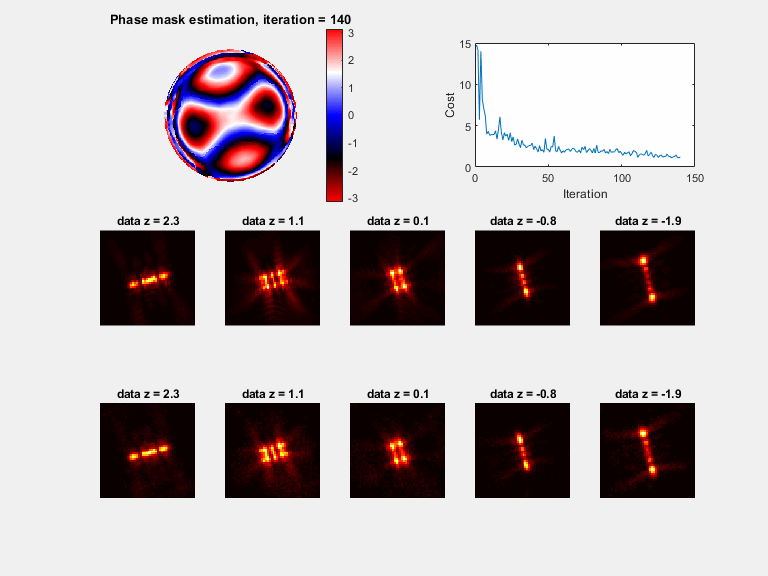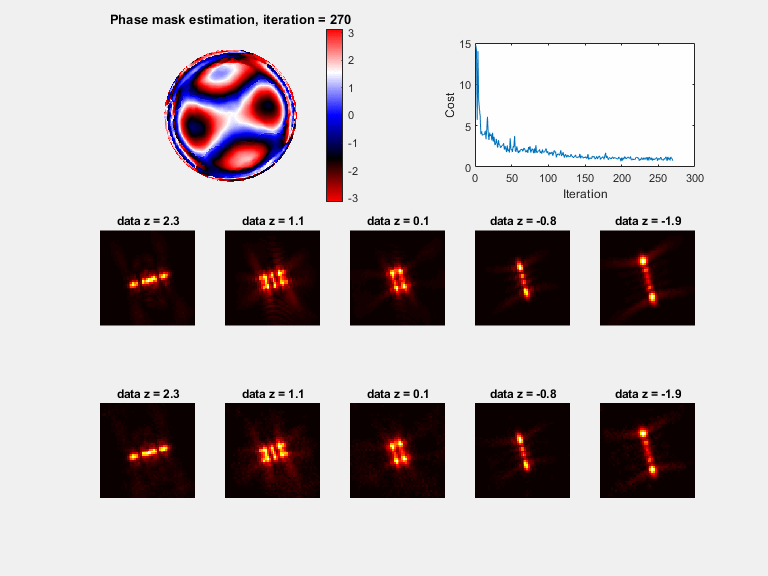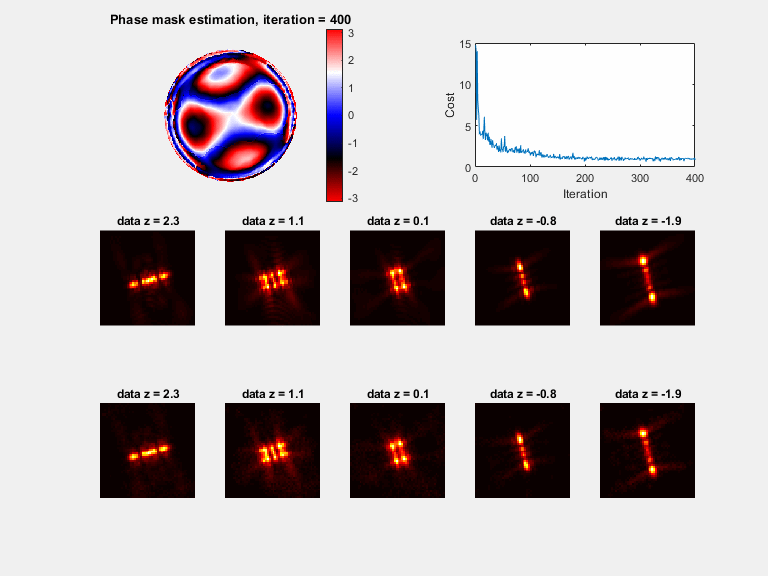 In microscopy, proper modeling of the image formation has a substantial effect on the precision and accuracy in localization experiments and facilitates the correction of aberrations in adaptive optics experiments. The observed images are subject to polarization effects, refractive index variations and system specific constraints. Previously reported techniques have addressed these challenges by using complicated calibration samples, computationally heavy numerical algorithms, and various mathematical simplifications. In this work, we present a phase retrieval approach based on an analytical derivation of the vectorial diffraction model. Our method produces an accurate estimate of the system phase information (without any prior knowledge) in under a minute.
In microscopy, proper modeling of the image formation has a substantial effect on the precision and accuracy in localization experiments and facilitates the correction of aberrations in adaptive optics experiments. The observed images are subject to polarization effects, refractive index variations and system specific constraints. Previously reported techniques have addressed these challenges by using complicated calibration samples, computationally heavy numerical algorithms, and various mathematical simplifications. In this work, we present a phase retrieval approach based on an analytical derivation of the vectorial diffraction model. Our method produces an accurate estimate of the system phase information (without any prior knowledge) in under a minute.
For more details:
B. Ferdman, E. Nehme, L. E. Weiss, R. Orange, O. Alalouf, Y. Shechtman “VIPR: Vectorial Implementation of Phase Retrieval for fast and accurate microscopic pixel-wise pupil estimation” bioRxiv (2020)
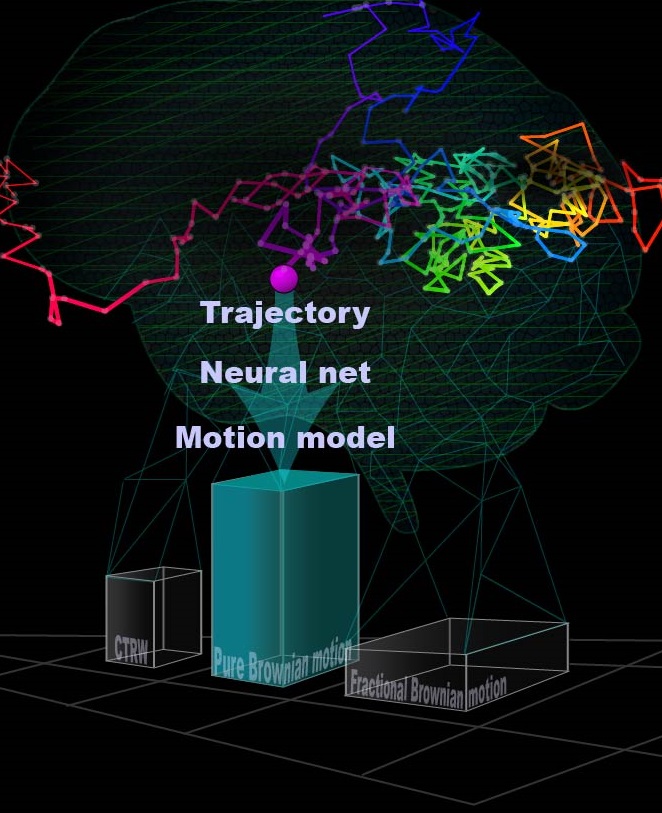 Diffusion plays a crucial role in many biological processes including signaling, cellular organization, transport mechanisms, and more. Direct observation of molecular movement by single-particle-tracking experiments has contributed to a growing body of evidence that many cellular systems do not exhibit classical Brownian motion, but rather anomalous diffusion. Characterization of the physical process underlying anomalous diffusion remains a challenging problem for several reasons. First, different physical processes can exist simultaneously in a system. Second, commonly used tools for distinguishing between these processes are based on asymptotic behavior, which is experimentally inaccessible in most cases. Finally, different transport modes can result in the same diffusion power-law α, typically obtained from the mean squared displacements (MSD).
Diffusion plays a crucial role in many biological processes including signaling, cellular organization, transport mechanisms, and more. Direct observation of molecular movement by single-particle-tracking experiments has contributed to a growing body of evidence that many cellular systems do not exhibit classical Brownian motion, but rather anomalous diffusion. Characterization of the physical process underlying anomalous diffusion remains a challenging problem for several reasons. First, different physical processes can exist simultaneously in a system. Second, commonly used tools for distinguishing between these processes are based on asymptotic behavior, which is experimentally inaccessible in most cases. Finally, different transport modes can result in the same diffusion power-law α, typically obtained from the mean squared displacements (MSD).
We implement a neural network to classify single-particle trajectories by diffusion type: Brownian motion, fractional Brownian motion (FBM) and Continuous Time Random Walk (CTRW). Furthermore, we demonstrate the applicability of our network architecture for estimating the Hurst exponent for FBM and the diffusion coefficient for Brownian motion on both simulated and experimental data. The networks achieve greater accuracy than MSD analysis on simulated trajectories while requiring as few as 25 steps. On experimental data, both net and MSD analysis converge to similar values, with the net requiring only half the number of trajectories required for MSD to achieve the same confidence interval.
For more details:
N. Granik, L. E. Weiss, E. Nehme, M. Levin, M. Chein, E. Perlson, Y. Roichman, Y. Shechtman “Single particle diffusion characterization by deep learning“, Biophysical Journal 117, 2,185-192 (2019).
 We present a refractometry approach in which the fluorophores are preattached to the bottom surface of a microfluidic channel, enabling highly-sensitive determination of the Refractive Index using tiny amounts of liquid by detecting the Supercritical Angle Fluorescence (SAF) effect at the conjugate back focal plane of a high NA -obejective.
We present a refractometry approach in which the fluorophores are preattached to the bottom surface of a microfluidic channel, enabling highly-sensitive determination of the Refractive Index using tiny amounts of liquid by detecting the Supercritical Angle Fluorescence (SAF) effect at the conjugate back focal plane of a high NA -obejective.
The SAF effect (presented above) is the propagation of evanescent waves in the higher refractive index immersion medium, which captures the change in the transfer coefficients, observed as a strong transition ring.
We demonstrate the relevance of our system for monitoring changes in biological systems. As a model system, we show that we can detect single bacteria (Escherichia coli) and measure population growth.

Left: White light images of bacterial growth. Right: Corresponding changes in the refractive index.
For more details:
B. Ferdman, L.E. Weiss, O. Alalouf, Y. Haimovich, Y. Shechtman “Ultrasensitive refractometry via supercritical angle fluorescence“, ACS Nano, 12, 11892-11898 (2018).
*** See also ACS Nano perspective (12/2018)
Deep learning has been shown to be an effective tool for image classification. Here we demonstrate that capability extends to distinguishing the colors of single emitters from grayscale images as well. This was done by training a convolutional neural network (CNN) on a library of images containing up to four types of quantum dots with different emission wavelengths embedded in a polymer matrix, then evaluating the net with new images. Surprisingly, we found that the same approach was applicable to the much more challenging problem of classifying moving emitters as well, where the chromatic-dependent subtleties in the point-spread function (PSF) are distorted by motion blur. The performance of the neural net in these two applications show that such an approach be used to simplify the design of multicolor microscopes by replacing hardware components with downstream software analysis. In a second application of neural nets, we have shown how a phase-modulating element, which can be used to control the shape of the PSF, can be designed in parallel with net training in order to optimize the ability of the net to distinguish the position and color of the object. This approach produces novel phase masks that increase the net’s ability to categorize emitters while maintaining other desirable properties, namely, the localizability of emitters. This approach for mask optimization solves a longstanding problem in PSF-engineering: how a phase mask can be optimally designed to encode for any parameter of interest. For more details: In localization microscopy, regions with a high density of overlapping emitters pose an algorithmic challenge. Various algorithms have been developed to handle overlapping PSFs, all of which suffer from two fundamental drawbacks: data-processing time and sample-dependent paramter tuning. Recently we demonstrated a precise, fast, parameter-free, superresolution image reconstruction by harnessing Deep-Learning. By exploiting the inherent additional information in blinking molecules, our method, dubbed Deep-STORM, creates a super-resolved image from the raw data directly. Deep-STORM is general and does not rely on any prior knowledge of the structure in the sample. For more details: How, and to what precision, can one determine the 3D position of a sub-wavelength particle by observing it through a microscope? This is the problem at the heart of methods such as single-particle-tracking and localization based super-resolution microscopy (e.g. PALM, STORM). One useful way of achieving such 3D localization at nanoscale precision is to modify the point-spread-function (PSF) of the microscope so that it encodes the 3D position in its shape. For more details: Our PSF optimization method can be used to generate PSFs with unprecedented capabilities, like an extremely large, modular axial (z) range of up to 20 um. The resulting optimal large-range PSFs belong to a family of PSFs we call the Tetrapod PSFs, and they look like this: We demonstrate experimentally the applicability of these Tetrapod PSFs in micro-fluidic flow profiling over a 20um z range, and in tracking under noisy biological conditions: Often in fluorescent microscopy one is interested in observing several different types of fluorescently labeled objects. Commonly, this is done by labeling different objects with different colors. How can you distinguish between different colors using a highly sensitive grayscale detector (e.g. EMCCD)? One approach is to separate the emission light into different color channels with dichroic elements. Alternatively, is possible to switch between emission filters and image sequentially. But how do you simultaneously image multiple colors on a single multiple channel with no additional elements (other than a 4F system with a phase mask)? The answer is multicolor PSF engineering. By exploiting the spectral response of the phase modulating element, it is possible to design masks that create different phase delays for different colors, and therefore enable simultaneous multicolor 3D tracking or multicolor super-resolution imaging. Red and green fluorescent microspheres diffusing in 3D in solution, imaged using a multicolor 20um Tetrapod PSF. The green PSF is designed to be rotated by 45 degress relative to the red PSF. Can you find the only green microsphere? For more details: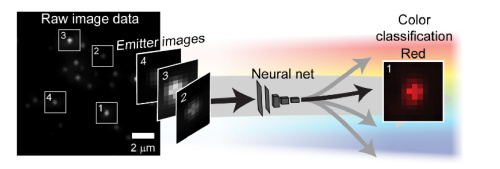 An illustration depicting a neural net decoding the color of objects from a grayscale image.
An illustration depicting a neural net decoding the color of objects from a grayscale image.
E. Hershko, L.E. Weiss, T. Michaeli, Y. Shechtman, “Multicolor localization microscopy and point-spread-function engineering by deep learning“, Optics Express 27, 5, 6158-6183 (2019)
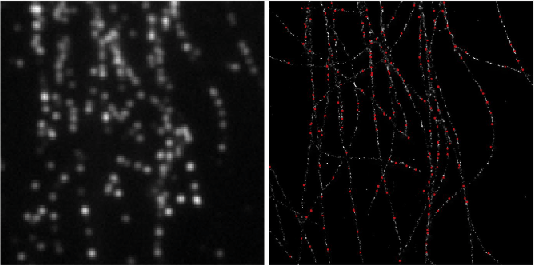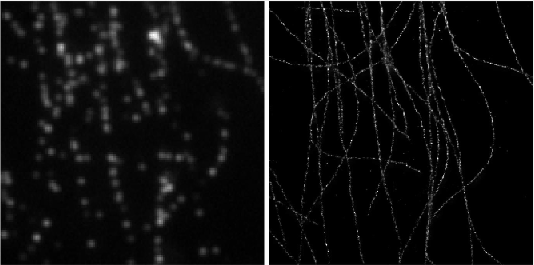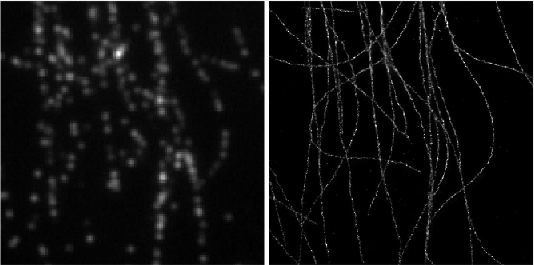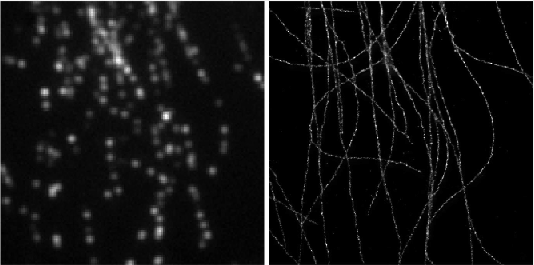
Left: Low resolution high density frames of microtubules labelled with Alexa 647. Right: The generated image by summing the super-resolved outputs from Deep-STORM.
E. Nehme, L.E. Weiss, T. Michaeli, and Y. Shechtman, “Deep-STORM: Super Resolution Single Molecule Microscopy by Deep Learning“, Optica 4, 458-464 (2018).
*** See also Nature Methods highlight ( May 2018)Optimal PSF for 3D localization microscopy
We have recently asked the basic question – what is the optimal way to modify a microscope’s PSF in order to encode the 3D position (x,y,z) of a point emitter in the most efficient way? We approach this challenge by solving an optimization problem: Find a pupil-plane phase pattern that yields a PSF which is maximally sensitive to small changes in the particle’s position. Formulated mathematically, this sensitivity corresponds to the Fisher-Information of the system. The result is the saddle-point PSF (bottom right panel in figure below):
Y. Shechtman, S.J. Sahl, A.S. Backer and W.E. Moerner, “Optimal point spread function design for 3D imaging“, Physical Review Letters 113, 133902 (2014).
Extremely-large-range PSFs for 3D localization microscopy

Y. Shechtman, L.E. Weiss, A.S. Backer, S.J. Sahl and W.E. Moerner, “Precise 3D scan-free multiple-particle tracking over large axial ranges with Tetrapod point spread functions“, Nano Letters,DOI:10.1021/acs.nanolett.5b01396 (2015).
Multicolor 3D PSFs
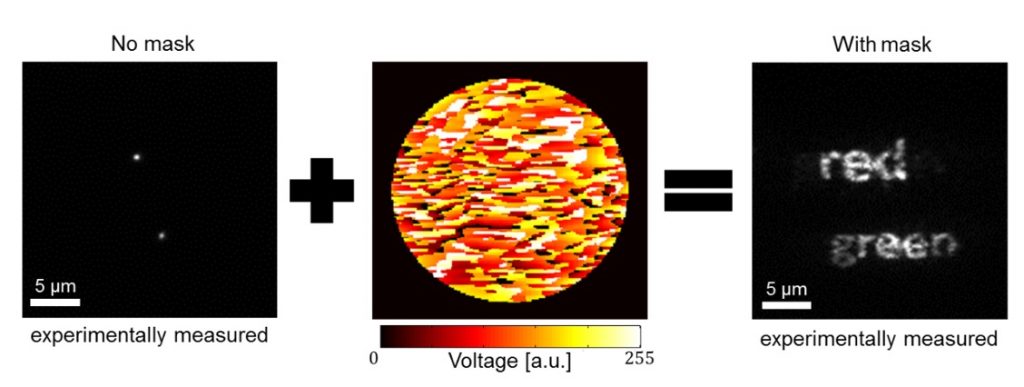

Y. Shechtman, L.E. Weiss, A.S. Backer, M.Y. Lee and W.E. Moerner, “Multicolor localization microscopy by point-spread-function engineering“,Nature Photonics 10, (2016).
*** See also Nature Methods highlight ( September 2016)

